A global collaboration led by the 4D Nucleome Consortium (4DN), under the guidance of Job Dekker, Ph.D., at UMass Chan Medical School, has produced groundbreaking models of the human genome’s three-dimensional structure. This study provides an unprecedented view of the genome’s architecture over time, revealing over 140,000 looping interactions between genes and their regulatory elements. The findings are published in the esteemed journal Nature.
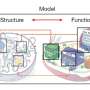
The research aims to link genomic structure with gene function by mapping the intricate folding of the human genome in both human embryonic stem cells and immortalized fibroblasts. By integrating data from more than a dozen advanced genomic and computational techniques, this project establishes a significant framework for future studies that investigate genome architecture and its impact on gene function.
Dr. Dekker, a Howard Hughes Medical Investigator and professor of systems biology, emphasized the importance of this mapping. He stated, “To understand how DNA translates linear genetic information into biologically meaningful actions, it’s essential to map the physical organization of the genome in space relative to itself and other structures in the nucleus.” The study’s map allows researchers to visualize how the genome is organized spatially and temporally, which is crucial for understanding its functional role.
The human genome comprises over 20,000 protein-coding genes and millions of regulatory elements that influence gene expression. Although many regulatory components have been cataloged, the mechanisms by which they interact with specific genes—especially across vast distances—remain largely elusive. The spatial organization of the genome significantly affects which regulatory elements and genes are in proximity, influencing their interaction.
The research uses an analogy of “cooked spaghetti” to illustrate how folding and looping mechanisms bring distant genomic elements closer together for interaction. Additionally, the genome’s structure can vary significantly from cell to cell and state to state, such as during cell division or stress responses, further affecting gene functionality.
Since its inception in 2015, the 4DN has focused on developing and refining experimental techniques to measure genomic structure in three dimensions. This latest study synthesizes efforts from over three dozen laboratories across eight countries, utilizing methodologies such as Hi-C, micro-C, SPRITE, ChiA-Pet, PLAC-seq, and GAM. Each technique provides unique insights into genomic folding and organization.
The study not only outlines how various experimental approaches contribute to understanding the genome but also offers a practical guide for researchers. It includes a decision tree that details the strengths of different methods, helping scientists choose the appropriate techniques for their specific research questions.
Through the integration of diverse datasets, the 4DN has compiled an extensive catalog of looping interactions that no single methodology could have detected independently. The research also initiates the assignment of dynamic genomic functions to multiple genes and regulatory elements within a three-dimensional framework. Furthermore, the data can be utilized to develop deep learning models that screen DNA sequences to uncover the mechanisms of genome folding and their relationship to gene function.
“This is the most detailed view of the living physical genome as it exists inside of cells,” said Dekker. “It’s the foundation for the deep exploration of structure and function of the genome.”
Looking ahead, the next phase of the 4DN project will focus on integrating genomic datasets with advanced imaging technologies to analyze nuclear changes during development and disease. This ongoing research promises to deepen our understanding of the human genome’s complexity and its implications for health and disease.